Batch Processing#
Cryo-electron tomography datasets frequently comprise tens to hundreds of tomograms, each requiring consistent processing. Manually applying the same operations to each tomogram is tedious and error-prone. This tutorial demonstrates how to construct reproducible workflows that process entire datasets automatically. Pipelines are useful for datasets of 10 or more tomograms, though the reproducibility benefits apply to smaller datasets as well. For individual samples, the GUI provides the same (or more) functionality with immediate visual feedback.
We use a Chlamydomonas reinhardtii dataset as our working example. Starting from membrane segmentations, we will generate seed points for constrained template matching of membrane-associated proteins.
Requirements#
Install Mosaic according to the installation instructions.
This tutorial assumes voxel-level membrane segmentations are available, with one file per tomogram:
segmentations/
├── tomo_001_seg.mrc
├── tomo_002_seg.mrc
├── ...
└── tomo_100_seg.mrc
Note
The batch processing functionality is available with version 1.1.0 of Mosaic.
Creating a Processing Pipeline#
Open the Pipeline Builder via File > Batch Processing (or Ctrl+Shift+P).
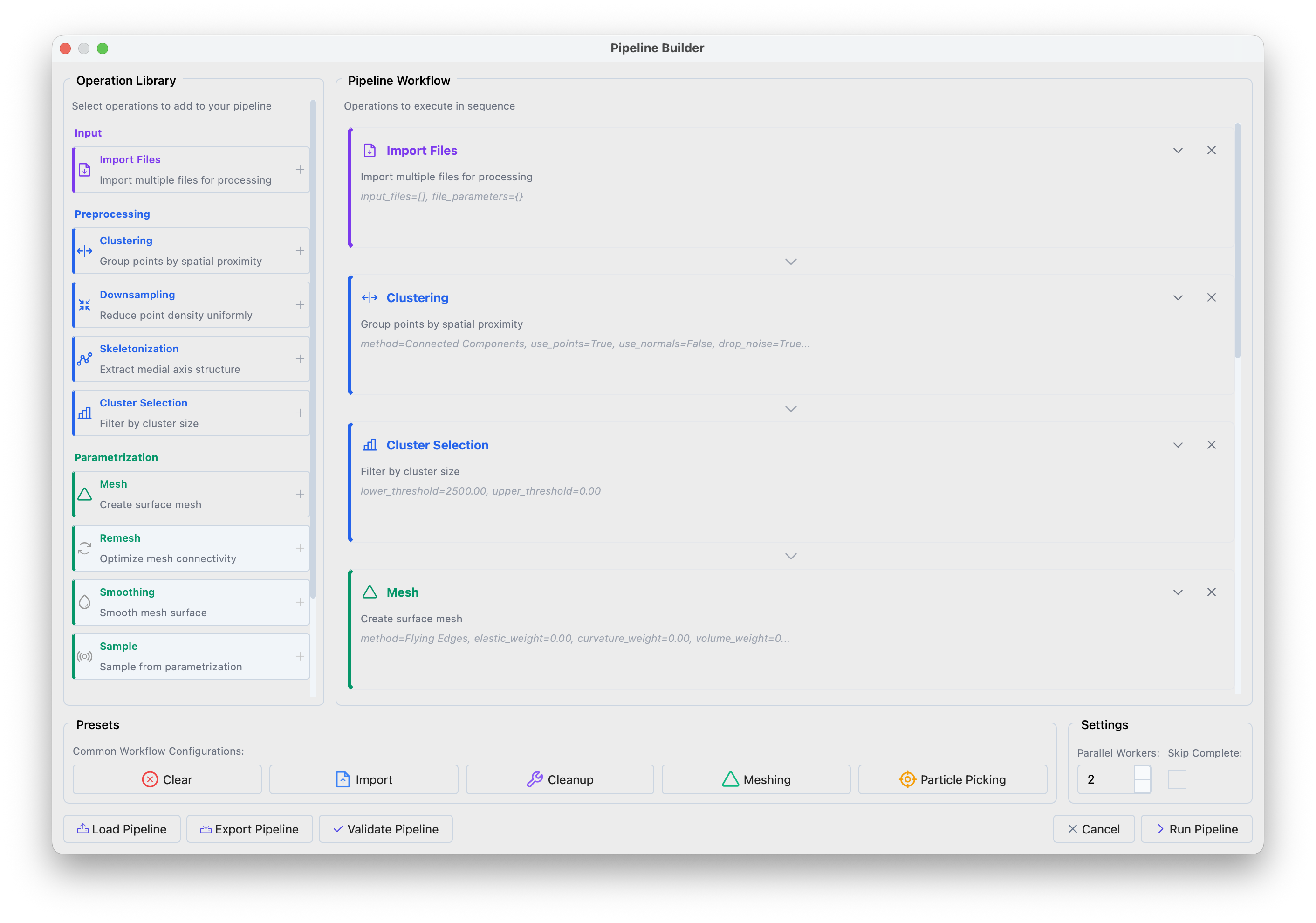
Pipeline Builder interface showing the operation library (left) and workflow panel (right).#
For seed point generation, click the Particle Picking preset in the bottom panel. This loads a standard workflow that we will customize for our data.
Configuring Input Files#
Expand the Import Files card by clicking on it, then click Select Files to choose your segmentation files. If your segmentations have incorrect file header information, click Configure Import Parameters to adapt them.
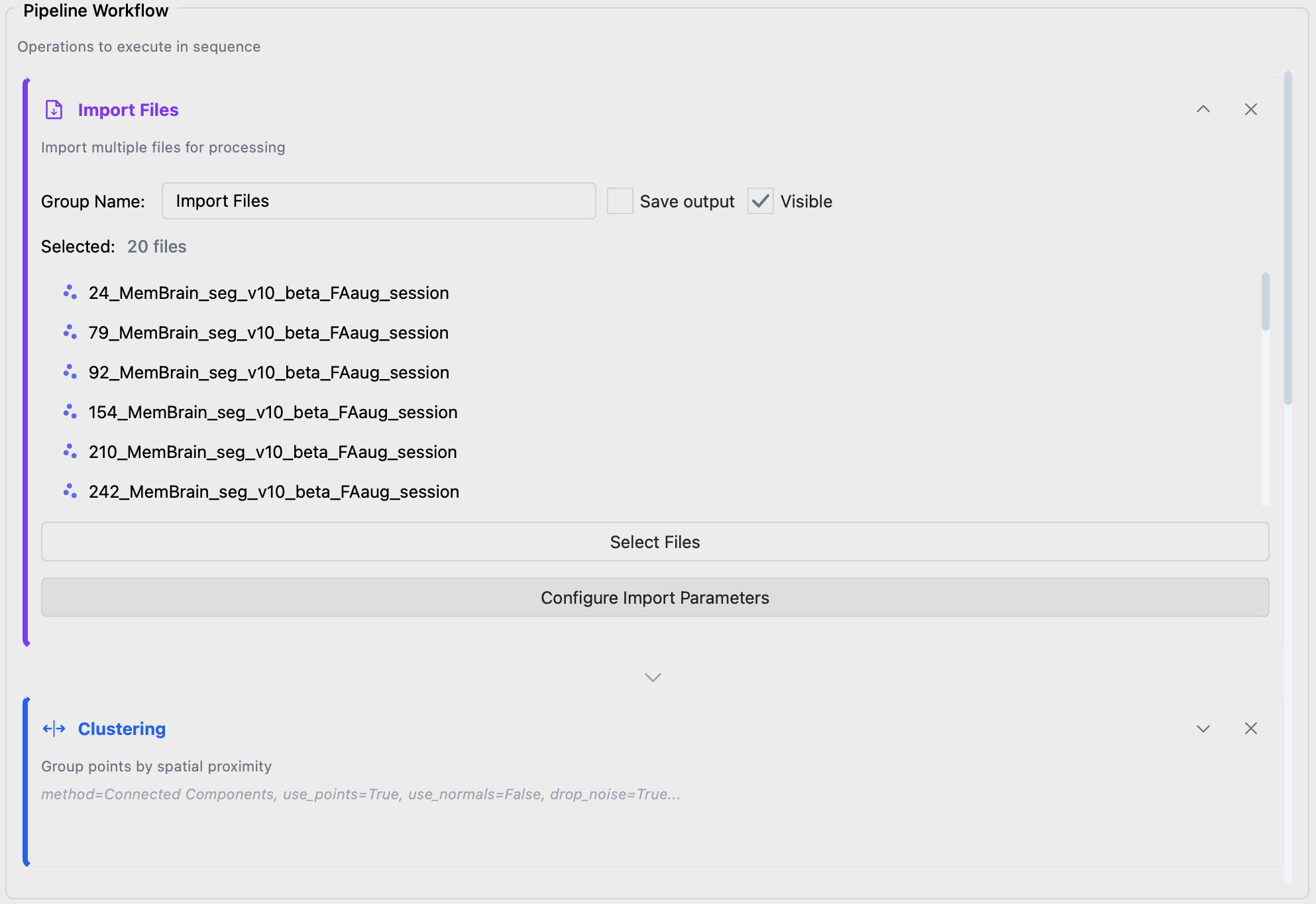
Import Files configuration.#
Understanding the Processing Steps#
Each operation card can be expanded to configure its parameters. The preset workflow performs the following transformations:
Clustering: Separates the segmentation into distinct membrane compartments
Filtering: Removes small fragments that are likely segmentation artifacts
Meshing + Smoothing: Creates a smooth triangular surface representation
Sampling: Places evenly-spaced points with surface normals along the membrane
Here we explain how to choose appropriate parameter values.
Clustering
Clustering separates your segmentation into distinct membrane compartments. A single segmentation file often contains multiple membranes (plasma membrane, ER, mitochondria, etc.) that should be processed independently.
Method: Connected Components identifies spatially separated regions
Distance: -1.0 uses single-voxel connectivity
Cluster Selection
Small clusters are typically segmentation artifacts rather than real membranes. This step removes them based on point count.
Lower Threshold: Minimum number of points to keep a cluster
The appropriate threshold depends on your tomogram’s pixel size and specimen. At 7Å/pixel, a threshold of 2000-8000 points is reasonable. At 14Å/pixel, the same physical membrane contains fewer voxels, so use a lower threshold (500-1000). Examine a few segmentations manually to determine what size range corresponds to real membranes versus noise.
Mesh
Meshing converts the point cloud into a triangular surface, enabling computation of surface normals.
Method: Flying Edges is fast and works well for initial mesh generation on dense volumes.
Meshes created with Flying Edges (and Marching Cubes) extract the contour of the segmentation, producing a closed surface that encloses the segmented region. This means seed points will be generated on both sides of the membrane, with normals pointing away from the enclosed volume on each side.
Alternative meshing methods (demonstrated in the Meshing pipeline preset) can fit a surface through the center of the segmentation rather than along its contour. These methods produce normals that all point in the same direction, which can be advantageous for template matching as it provides more precise spatial constraints, i.e., you search only on the biologically relevant side of the membrane rather than both sides.
Remesh and Smoothing
These steps create a cleaner surface representation. The original segmentation contains voxel-level noise that would result in inaccurate surface normals. Smoothing produces normals that better represent the true membrane orientation.
Smoothing Method: Taubin preserves surface shape while reducing noise
Iterations: 10 is typically sufficient
Sample
Sampling places seed points along the smoothed surface at regular intervals. These points, along with their surface normals, constrain downstream template matching to search only near the membrane surface.
Method: Distance ensures uniform spacing between points
Sampling: Distance between seed points in Angstroms. Choose a value smaller than the expected protein spacing, typically one-half to one-third the inter-protein distance. 30Å works well for most applications.
Offset: Distance from the surface toward the protein center. Set this to approximately half the height of your target protein.
Export Data
Format: STAR for compatibility with PyTME
Output Directory: Where to save the seed point files
Save Session
Saving sessions allows you to review results in Mosaic and make manual corrections if needed.
Output Directory: Where to save Mosaic session files
Execution Settings#
At the bottom of the Pipeline Builder:
Parallel Workers: Number of files to process simultaneously. Each worker requires approximately 8GB of memory.
Skip Complete: Skip files that already have output. Enable this to resume interrupted processing or re-run after adjusting parameters.
Running the Pipeline#
Click Run Pipeline to start processing. The status indicator in the bottom-right corner shows progress. For a typical workflow, expect 2-4 minutes per tomogram, scaling linearly with file count.
Upon completion, the Batch Navigator opens automatically.
Cluster Execution#
For large datasets or limited local resources, run pipelines on an HPC cluster:
Click Export Pipeline and save as
pipeline.jsonTransfer the configuration and data to your cluster
The mosaic-pipeline command executes pipelines from the command line:
# Process all files with 8 parallel workers
mosaic-pipeline pipeline.json --workers 8
# Resume processing, skipping completed files
mosaic-pipeline pipeline.json --workers 8 --skip-complete
# Preview runs without executing
mosaic-pipeline pipeline.json --dry-run
For job array submission on SLURM, save this script and run it as sbatch run_pipeline.sh:
#!/bin/bash
#SBATCH --job-name=mosaic_batch
#SBATCH --cpus-per-task=2
#SBATCH --mem=12G
#SBATCH --time=01:00:00
#SBATCH --output=logs/mosaic_%A_%a.out
CONFIG="/path/to/pipeline.json"
# Self-submitting: first run determines array size
if [ -z "$SLURM_ARRAY_TASK_ID" ]; then
mkdir -p logs
N_RUNS=$(mosaic-pipeline "$CONFIG" --dry-run | head -1 | grep -oP '\d+')
sbatch --array=0-$((N_RUNS-1)) "$0"
exit 0
fi
mosaic-pipeline "$CONFIG" --index $SLURM_ARRAY_TASK_ID
The script automatically determines the number of input files and submits itself as a job array.
Reviewing Results#
The Batch Navigator enables efficient inspection of processed sessions. You can open it via File > Batch Navigator (or Ctrl+Shift+N)
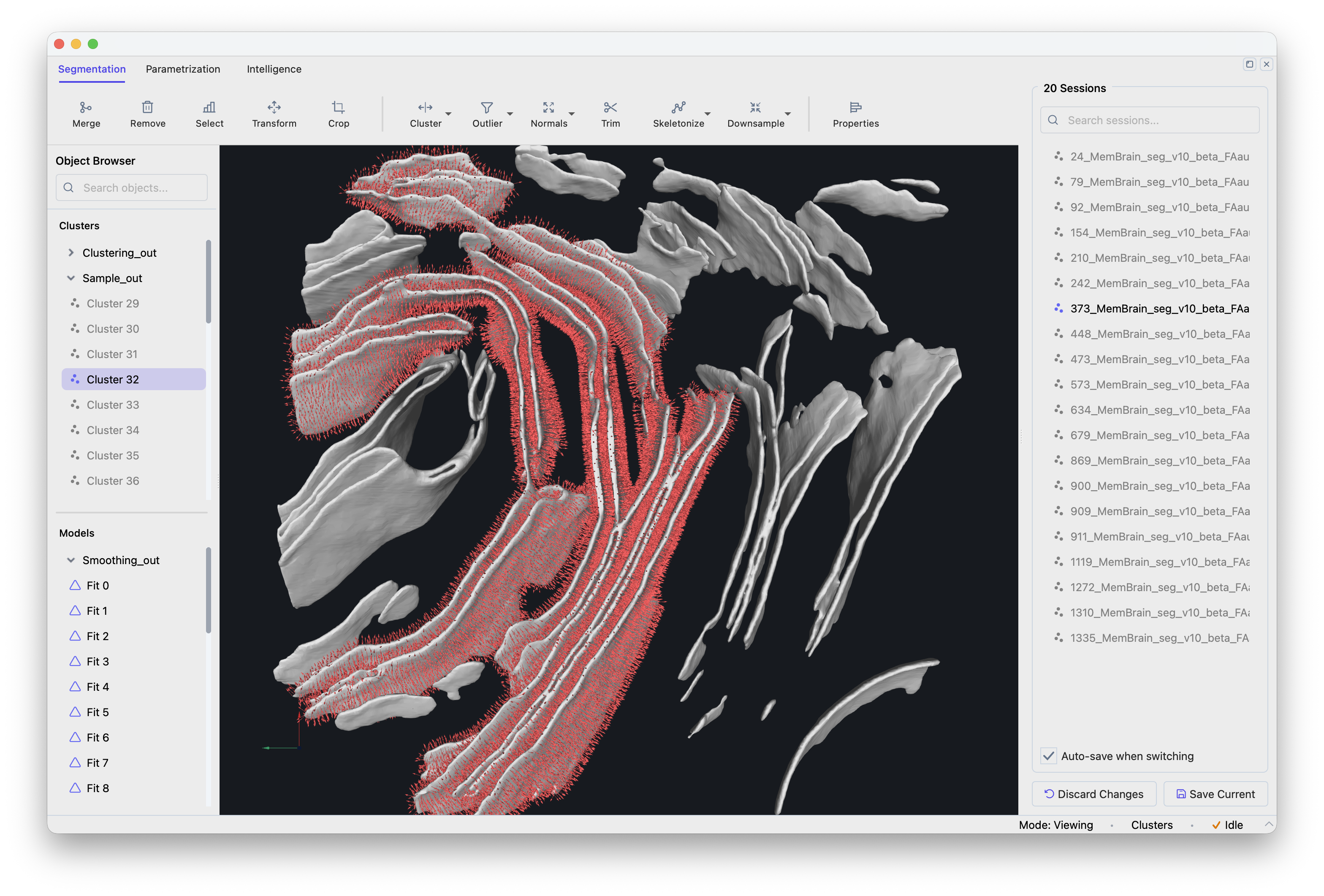
Mosaic interface with batch Navigator on the right showing the session list.#
Click any session to load it. The navigator auto-saves modifications when switching sessions.
For quality control, verify:
Distinct membranes are properly separated
Small artifacts were filtered out
The mesh surface follows the membrane smoothly
Seed points are evenly distributed with consistent orientations
If a session needs correction, use Mosaic’s selection and editing tools to adjust. Changes are saved automatically when you switch to another session. The remaining buttons can be used to discard changes to the current session or to directly save it to disk.
Using Seed Points for Template Matching#
The exported STAR files contain seed point coordinates and surface normals for constrained template matching. In PyTME, configure via Intelligence > Template Matching > Setup, specifying your seed points file and search constraints (rotational uncertainty typically 15-20 degrees, translational uncertainty 5-10Å).
This provides an example configuration on how to setup constrained template matching. Have a look at the PyTME documentation on how to scale to the entire dataset.
Pipeline Operations Reference#
This section details all available operations for constructing custom pipelines.
Input#
- Import Files
Loads files for batch processing. Supported formats: Mosaic sessions (.pickle), point clouds (STAR, XYZ, TSV), meshes (OBJ, STL, PLY), and volumes (MRC, EM).
Preprocessing#
- Clustering
Partitions point clouds into spatially coherent groups.
Connected Components: Labels disjoint regions based on spatial connectivity. Distance parameter controls the connectivity threshold.
Leiden: Graph-based community detection using modularity optimization. Resolution parameter controls cluster granularity.
K-Means: Partitions into k clusters by minimizing within-cluster variance.
DBSCAN: Density-based clustering that identifies regions of high point density separated by low-density regions.
- Downsampling
Reduces point cloud density.
Radius: Removes points within a specified radius of each other while preserving original point positions.
- Skeletonization
Extracts the medial axis of tubular structures.
- Cluster Selection
Filters clusters by point count. Lower threshold removes clusters with fewer points; upper threshold removes clusters exceeding the specified count.
Parametrization#
- Mesh
Generates triangular surface meshes from point clouds.
Flying Edges: Fast marching cubes variant for isosurface extraction.
Poisson: Solves a Poisson equation to produce smooth, watertight surfaces. Requires consistent normal orientation.
Ball Pivoting: Reconstructs surfaces by rolling a ball of specified radius over the point cloud. Suitable for partial surfaces.
Alpha Shape: Computes the alpha complex, a generalization of the convex hull. Alpha parameter controls surface detail.
- Remesh
Modifies mesh topology.
Decimation: Reduces triangle count while preserving surface geometry. Reduction factor specifies the target reduction ratio.
Subdivision: Increases mesh resolution by subdividing triangles.
- Smoothing
Applies surface smoothing filters.
Taubin: Two-step filter that smooths iteratively without net shrinkage.
Laplacian: Iteratively moves vertices toward neighbor centroids. May cause mesh shrinkage.
- Sample
Generates point samples from mesh surfaces.
Distance: Places points at uniform spacing along the surface. Returns points with associated surface normals.
Points: Generates a specified number of uniformly distributed random samples.
Export#
- Export Data
Writes data to specified format.
Point clouds: STAR, XYZ, TSV
Meshes: OBJ, STL, PLY
Volumes: MRC, EM, H5
- Save Session
Serializes the complete Mosaic session state.
Troubleshooting#
- Memory errors during parallel execution
Reduce worker count. Each worker requires approximately 8GB RAM depending on segmentation size and number of intermediate outputs stored.