Influenza A Virus#
This tutorial guides you through analyzing Influenza A Virus (IAV) virus-like particles (VLPs) using Mosaic, taking you from initial segmentation to creating coarse-grained Martini models.
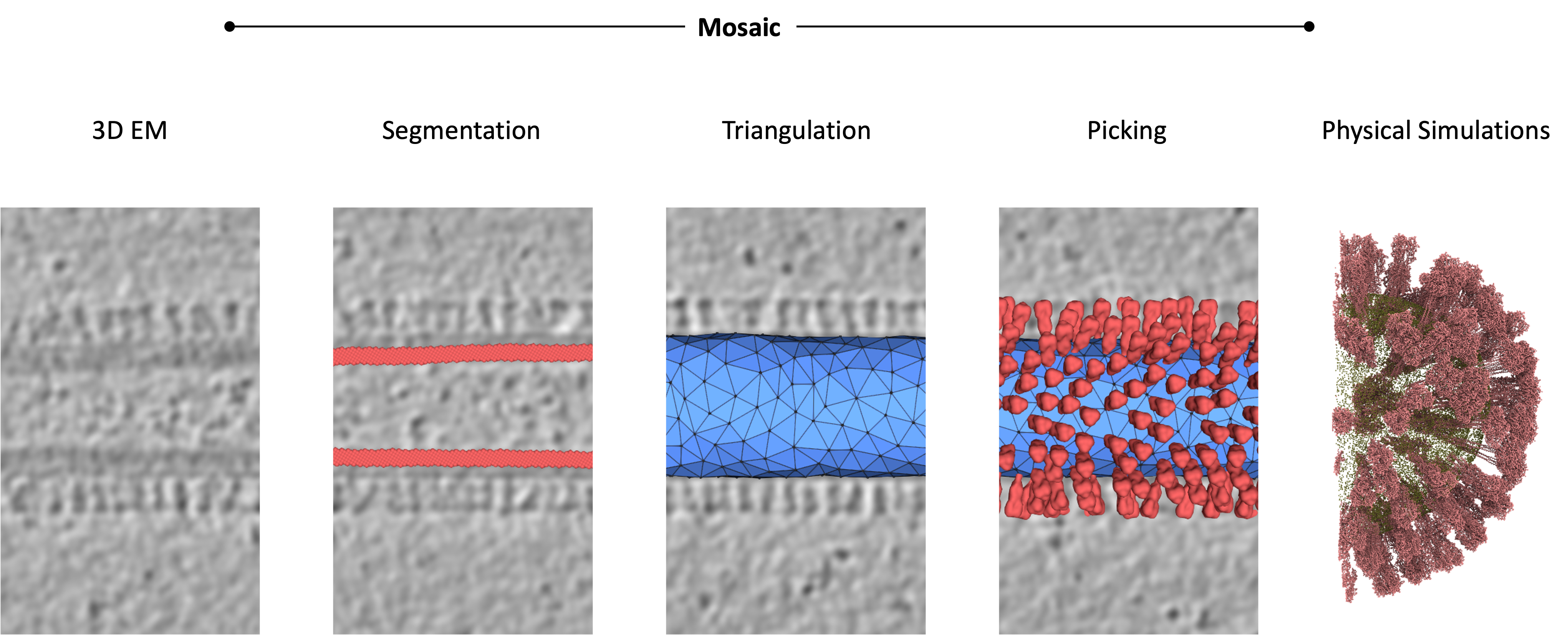
Coming up#
Requirements#
First, install Mosaic according to the installation instructions. For HMFF functionality, install the additional requirements listed in DTS Simulations. If you plan to backmap DTS models to coarse-grained representations, install the tools listed in the DTS Backmapping section.
Note
Membrane segmentation, template-matching and the equilibration of coarse-grained model equilibration require a GPU to complete in reasonable time. You can download intermediate results of compute-intensive task and additional material from ownCloud.
We’ll use publicly available cryo-ET data of an IAV VLP [1], which you can download from EMDB-11075 or via command line
wget https://ftp.ebi.ac.uk/pub/databases/emdb/structures/EMD-11075/map/emd_11075.map.gz
gunzip emd_11075.map.gz && mv emd_11075.map emd_11075.mrc
For membrane segmentation, download the MemBrain_seg_v10_alpha weights [2] from Google Drive.
Visual Demonstration#
The video below demonstrates the workflow from raw cryo-ET data to initial meshes, covering membrane segmentation, mesh generation, and refinement steps detailed in the following sections.
Membrane Segmentation#
Launch Mosaic and navigate to the Intelligence tab.
Click on the arrow next to the Membrane button and configure:
Click the Browse button to select the downloaded model ckpt file.
Window Size: 160
Clustering: Enabled
Augmentation: Enabled
Output Sampling: 12.0
Click Apply and select the downloaded IAV VLP tomogram.
The status indicator will change from “Ready” to “Membrane Segmentation.” The results will automatically load into Mosaic when complete.
Note
Membrane segmentation requires a GPU. If unavailable, download the pre-computed segmentation emd_11075_MemBrain_seg_v10_alpha.ckpt_segmented.mrc.gz and load it using File > Open.
Mesh Creation#
Clean the Segmentation#
Switch to the Segmentation tab
Remove clusters corresponding to small artifacts
In the Object browser, select the Cluster section.
Click the Select button from the Base operations menu, a window should open.
Use the slider at the bottom of the window to adjust the threshold for cluster selection. To remove small artifacts, drag the right slider to the left until only the central IAV VLP remains unselected.
Close the window, the selected clusters should now be highlighted in the Object browser as well in the viewer.
Press delete key or use Remove menu button to remove the selected clusters.
Click the Select button from the Base operations menu, a window should open.
Use the slider at the bottom of the window to adjust the threshold for cluster selection. To remove the small artifacts, drag the right slider to the left until only the central IAV VLP remains unselected.
Close the window, the selected clusters should now be highlighted in the Object browser as well in the viewer.
Press delete key or use Remove menu button to remove the selected clusters
Eliminate incorrectly segmented voxels using manual selection
Press r key - the mouse cursor should change to a crosshair
Click and drag to select the incorrectly segmented voxels at one end of the IAV VLP near the VLP end at the edge of the tomogram.
Press delete key or use Remove menu button to remove the selected voxels
Thin the segmentation.
Select the central IAV VLP in the Object Browser.
In the Segmentation tab, click the arrow next to the Thin button.
Choose the outer option to extract the outer segmentation layer.
Click Apply to apply the operation. A new object will appear in the Model section of the Object Browser.
Righ click on the previous object and click hide to clearly see the result of the thinning operation.
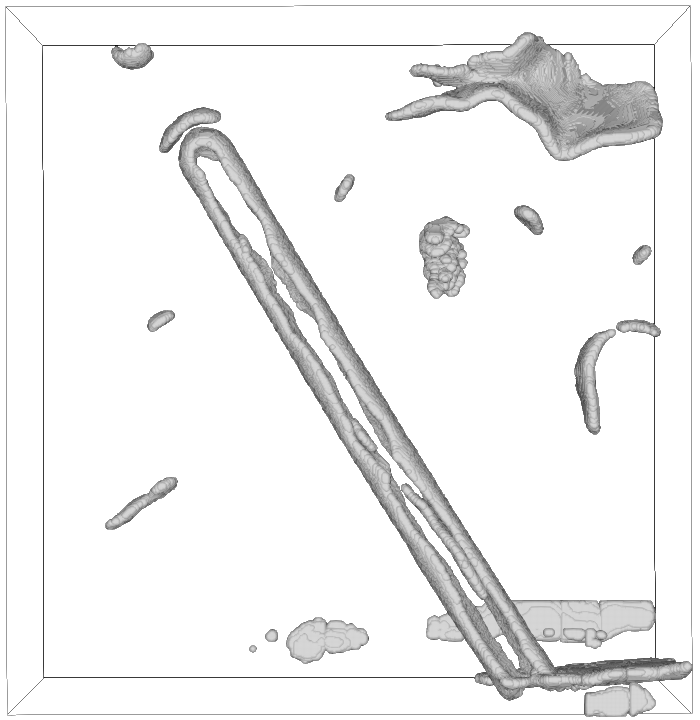
Before segmentation cleanup
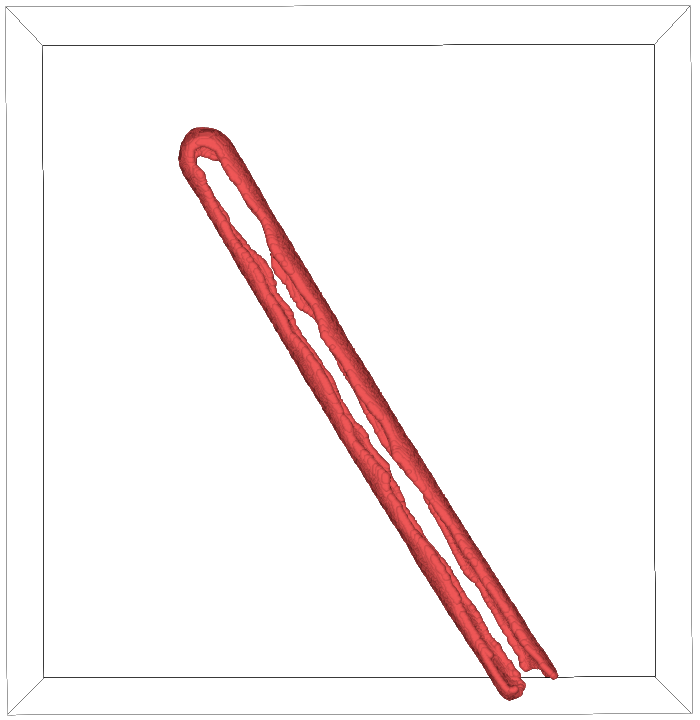
After segmentation cleanup
Generate Initial Mesh#
Switch to the Parametrization tab.
Select the cleaned segmentation in the Cluster section of the Object Browser.
Click on the arrow next to the Mesh buton and configure:
Method: Ball Pivoting
Elastic Weight: 1.0
Curvature Weight: 10.0
Volume Weight: 0.0
Boundary Ring: 0
Neighbors: 15
Radii: 60.0
Hole Size: -1.0
Downsample: True
Smoothing Steps: 5
Click Apply to fit the mesh, creating a new object in the Model section of the Object Browser.
Right-click the new mesh object and select Representation to change its visualization to Mesh for better clarity.
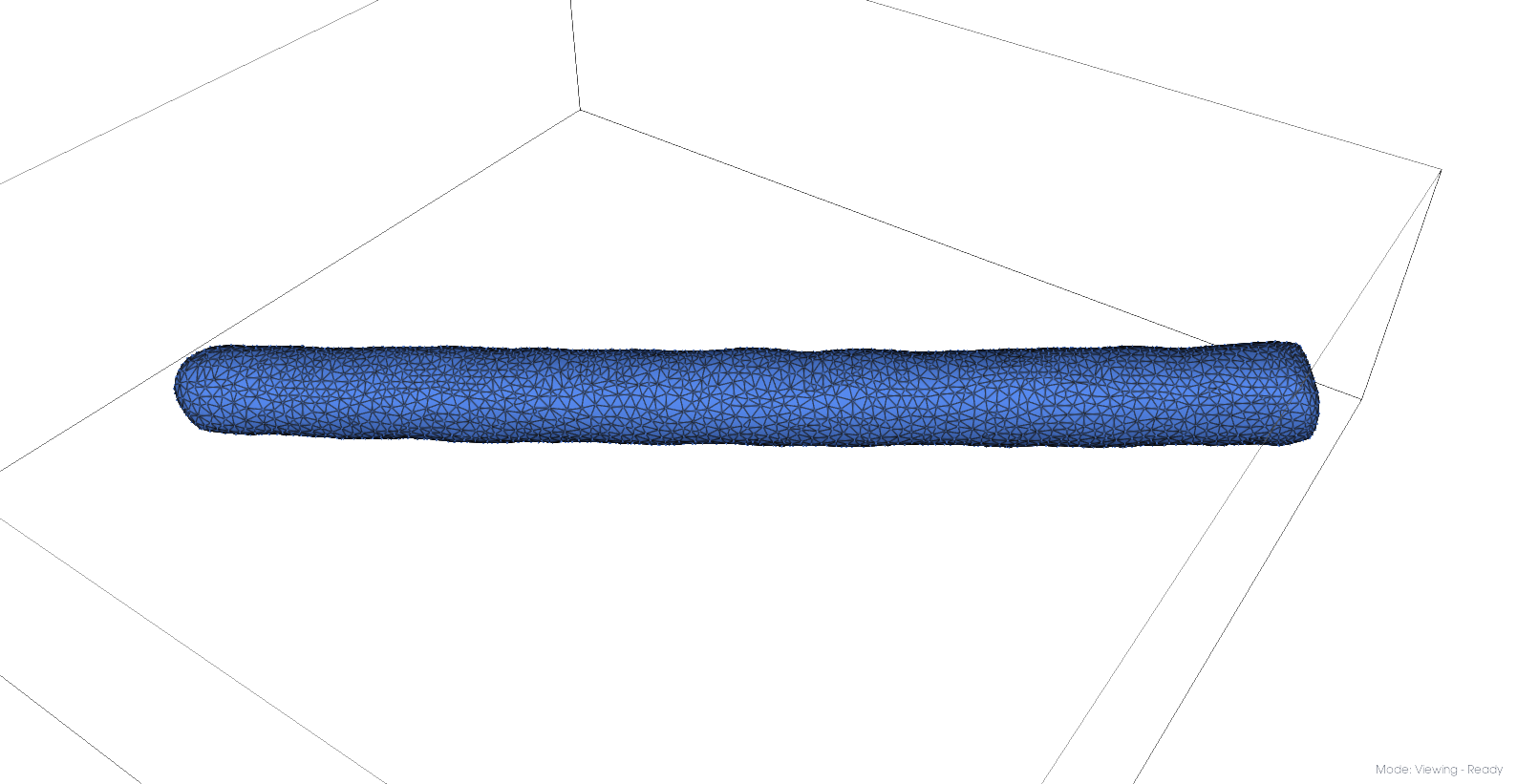
Initial mesh#
Refine the Mesh#
Since one cap of the IAV VLP falls outside the tomogram, and we’ll fill the mesh in that region to mitigate boundary effects in subsequent simulations:
Sample points from the created mesh.
Select the mesh in the Object Browser.
Click on the arrow next to the Sample button and set:
Sampling Method: Points
Sampling: 30000
Click Apply.
Create a new mesh from the cleaned samples.
Select the cleaned samples in the Object Browser.
Click the arrow next to Mesh again and configure:
Method: Ball Pivoting
Elastic Weight: 1.0
Curvature Weight: 10.0
Volume Weight: 0.005
Boundary Ring: 0
Neighbors: 15
Radii: 60.0
Hole Size: -1.0
Downsample: True
Smoothing Steps: 5
Click Apply to fit the mesh.
Right-click the new mesh object and select Representation to change its visualization to Mesh for better clarity. Hide the segmentation object to see the mesh clearly and compare it with the original mesh.
Note
Observe the filled cap of the IAV VLP, which extends beyond the original segmentation.
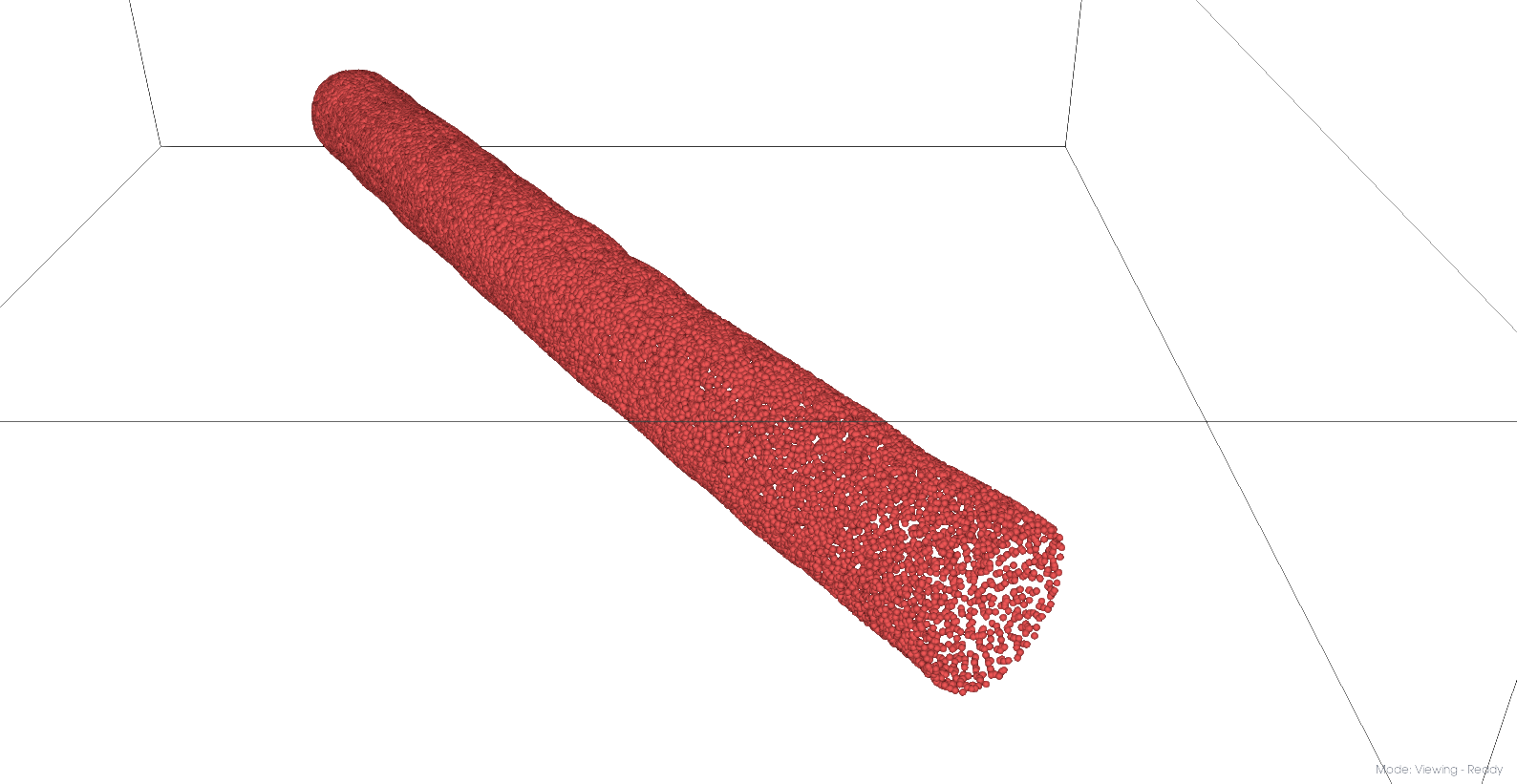
Cleaned mesh points
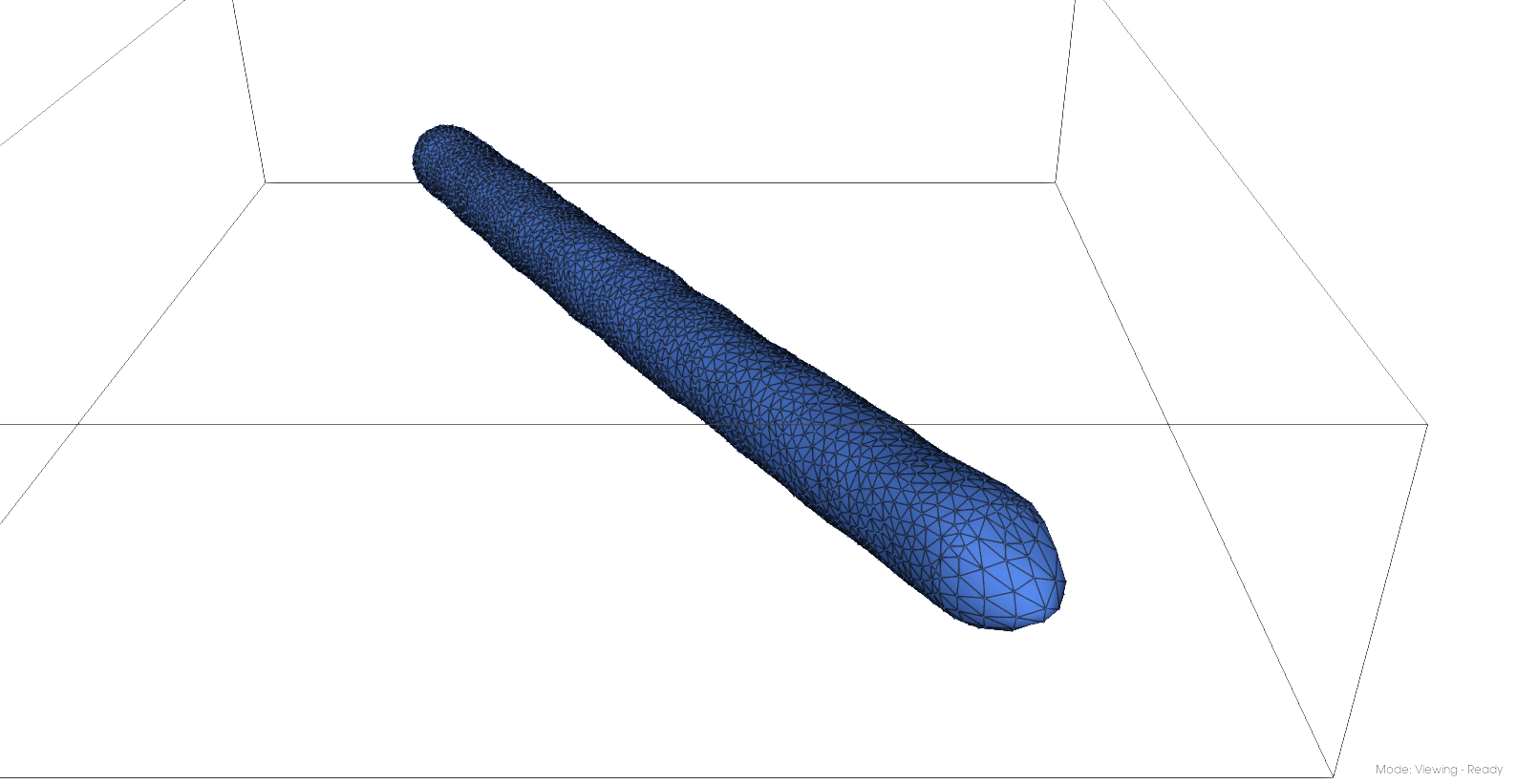
Pressurized mesh
Equilibrate the Mesh#
Before DTS simulation, meshes require equilibration to ensure stability and physical validity:
Select the refined mesh model from the Object Browser
Navigate to the Intelligence tab and click Equilibrate
Configure:
Average Edge Length: 100
Steps: 5000
Other parameters at default values
Once complete, Mosaic will create three meshes in the target directory:
mesh_base: the input mesh
mesh_remeshed: input mesh with desired edge length
mesh_equilibrated: the fully equilibrated mesh using Trimem [3]
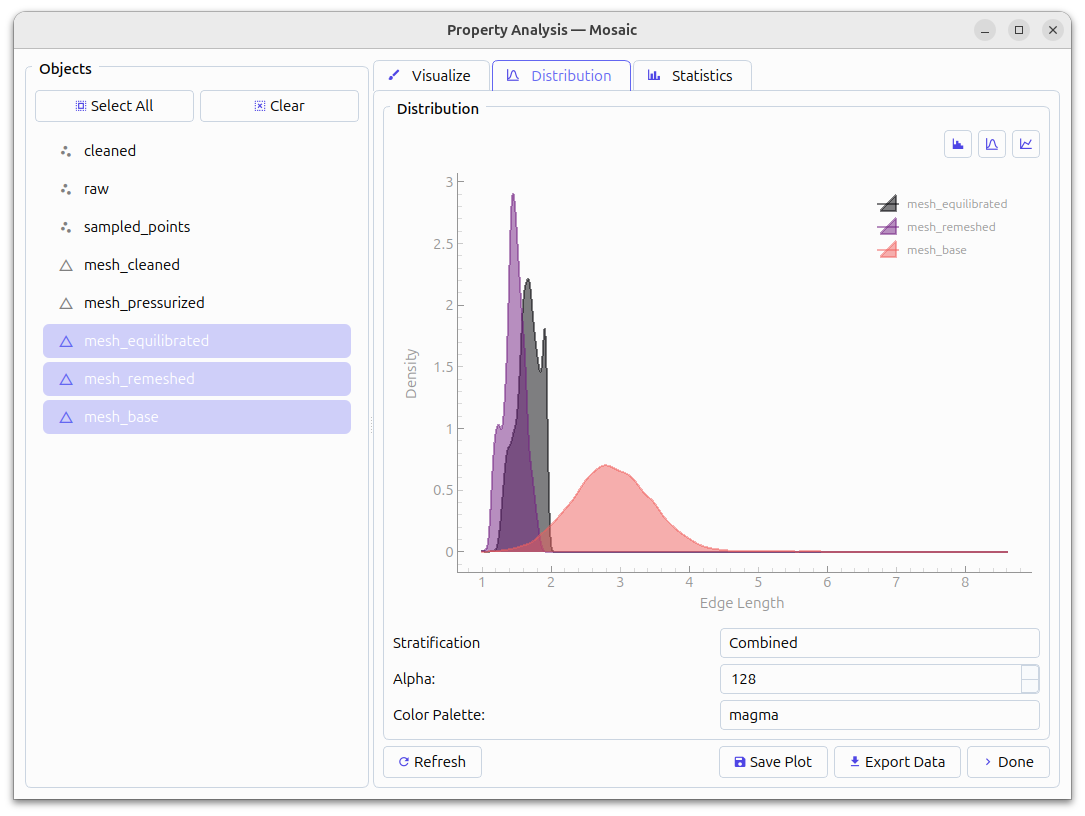
Comparison of edge lengths#
To assess edge-length distribution, import the meshes into Mosaic and use the Properties button in the Segmentation tab. We typically choose the equilibrated mesh for DTS simulation due to its smoother surface and more predictable behavior.
Note
Pre-computed equilibrated meshes are available from ownCloud.
HMFF Simulation#
Navigate to the Intelligence tab and click Setup in the DTS Simulation section. Configure:
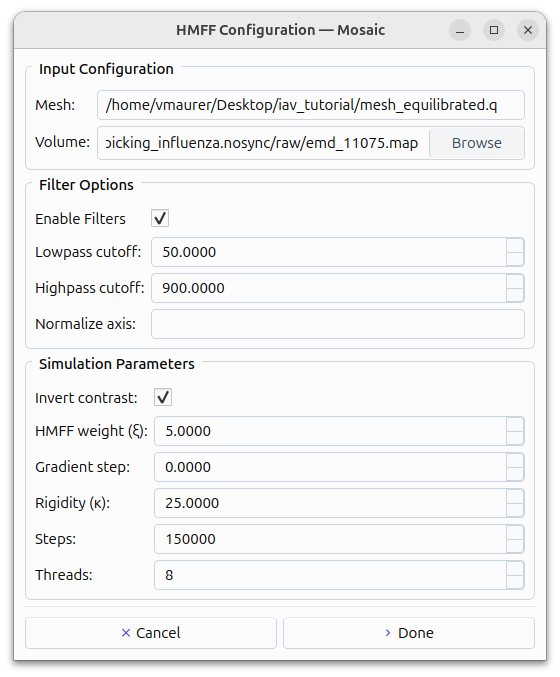
HMFF simulation setup dialog#
Mesh: Select mesh_equilibrated.q
Volume: Select the downloaded EMD-11075.
Invert Contrast: Enabled
HMFF weight (ξ): 5.0
Rigidity (κ): 25.0
Steps: 150000
Threads: 8 (or 1 for Mac unless FreeDTS is properly configured)
Lowpass cutoff: 50Å
Highpass cutoff: 900Å
This will create a filtered density map and setup files for DTS simulation [4] with HMFF. Open input.dts and set:
AlexanderMove = MetropolisAlgorithmOpenMP 0
VolumeCoupling = SecondOrder 0.6 1000 1.1
Run the simulation (takes less than five minutes with 8 threads):
bash run.sh
Note
Simulation outputs are available on ownCloud in hmff/TrajTSI_Done.
To analyze the refined mesh in Mosaic:
Click the arrow next to the Trajectory button in the Intelligence tab
Configure the settings to match the input.dts file:
Scale: 0.012202743213335199
Offset: 21.0,6.0,16.0
Mosaic will load all trajectory time points. Use View > Trajectory player to navigate through them. To assess the results, open the density file in View > Volume Viewer and adjust contrast as needed. Compare the mesh at step 0 (left) and step 150,000 (right) to see how HMFF has refined the mesh to better match the viral membrane.

Initial mesh.
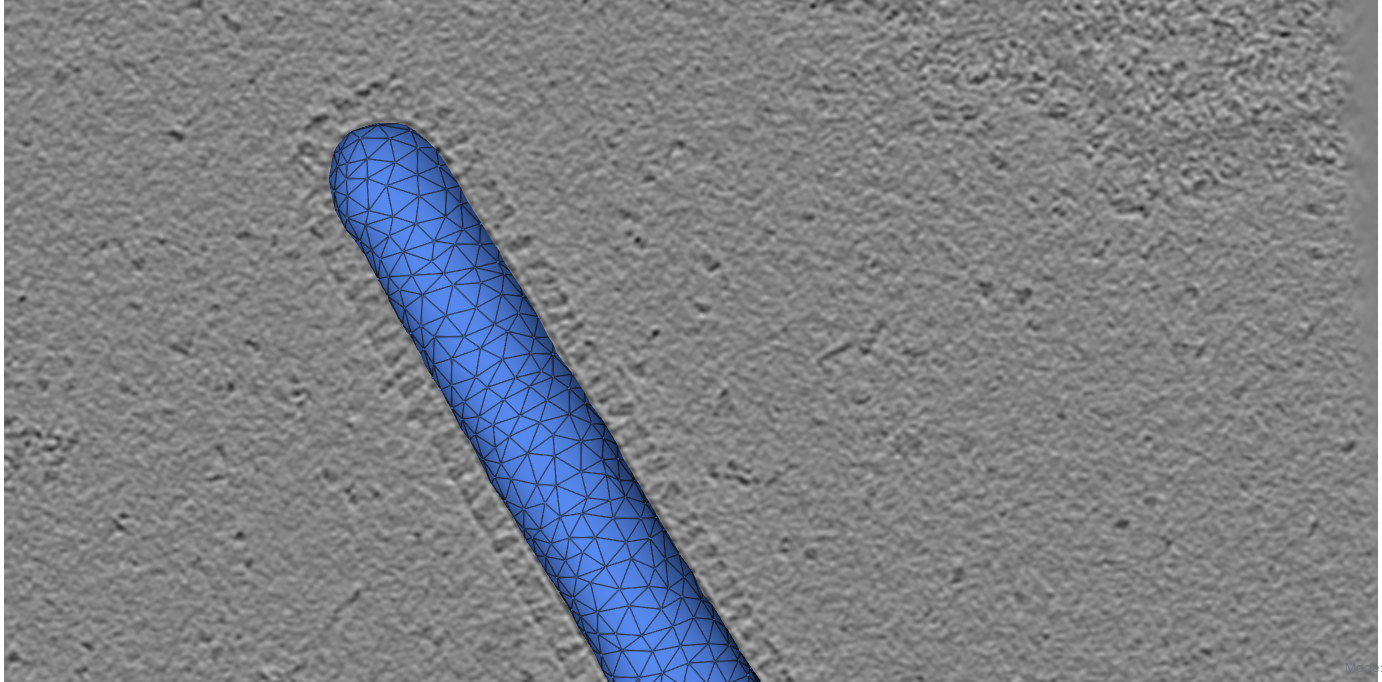
HMFF-refined mesh.
Note
If you notice vertices frozen in place throughout the simulation, this indicates the simulation is unable to develop them. Try increasing Min_Max_Lenghts or choose a lower edge length for equilibration to increase mesh resolution.
Constrained Template Matching#
Generate Seed Points#
To create seed points from the HMFF-refined mesh:
Select your desired time-point in the trajectory
Right-click the trajectory object in the Object Browser and select Duplicate
Move to the Parametrization tab and configure Sample:
Sampling Method: Distance
Sampling: 40
Offset: 100
This generates seed points approximately 40Å apart with a 100Å offset from the surface, which should position them near the centers of HA and NA proteins. Both can be validated using the Properties button in the Analysis section of the Segmentation tab. The offset should roughly correspond to the center of the protein-of-interest, in our case Hemagglutinin (HA) and Neuraminidase (NA).
Export the cluster object as a STAR file by right-clicking on it.
Template Matching#
The following outlines how to perform constrained template matching using PyTME [5].
Launch the PyTME Template Matching Dialog:
Navigate to the Intelligence tab
Click on Setup in the Template Matching directive.
Prepare Data: - In the “Data” tab, specify your working directory - Set paths to the EMD-11075 tomogram and the HA/NA structures
Prepare Templates:
Switch to the “Preprocess” tab to configure template preparation
Set Lowpass to 15
Set Align Template Axis to z
Set Flip Template to checked
Configure Template Matching:
In the “Matching” tab configure template matching parameters.
Set Angular Step to 7
Set Score Function to FLC
Set the path to the STAR file with seed points
Set Rotational Uncertainty to 15
Set Translational Uncertainty to (6,6,10) for HA and (6,6,12) for NA due to the longer stalk.
Set Tilt Range to 60,60 to create a wedge mask from -60 to 60 degrees.
Set Wedge Axes to 2, 0
Set Defocus to 30000
Set No Centering to checked
Set Peak Calling Parameters:
Switch to the “Peak Calling” tab
Set Peak Caller PeakCallerMaximumFilter
Set Number of Peaks 10000
Set Minimum Distance 7 for HA and 10 for NA
Configure Compute Resources:
In the “Compute” tab, allocate CPU cores and memory
Set backend cupy.
Execute the Workflow:
Click “OK” to generate the template matching scripts
Mosaic will create and organize all necessary files in your working directory
Run the generated scripts to perform template matching
Note
Template matching results are available from ownCloud.
Refine Protein Picks#
The template matching process generates coordinate files for HA and NA that need filtering:
Keep the top 97% of NA picks by score.
Visualize and manually refine the picks in Mosaic using the selection tool (or the GUI provided with PyTME).
Remove HA picks that are within 7 voxels of NA picks to avoid clashes.
Example filtering scripts are available from ownCloud, namely pytme/filter.py and pytme/resolve_clash.py.
Coarse-Grained Martini Models#
Backmapping#
We can now combined HMFF-refined membrane models with experimentally determined protein positions to create coarse-grained Martini model representations of IAV VLP
In the Intelligence tab, click Backmapping and select an output directory
Set target edge length to 20 (corresponds to 20Å in this case) and add both NA and HA inclusions
Navigate to the specified directory
Tip
The output directory will also contain a file mesh.tsi, which can be used for equilibrium DTS simulations with protein inclusions.
Coarse Graining#
Open the file martinize.sh and add paths to PDB files for NA_STRUCTURE and HA_STRUCTURE. Save the file and run
bash martinize.sh
Note
The principal axis of both proteins is required to align with the z-axis. This can be achieved with different tools. An example script using PyTME is available from ownCloud, namely pytme/templates/rot_structures.py.
Once the command above is completed, we can create a coarse-grained representation of the entire system using TS2CG [6]
# Use PLM utility to create a bilayer
bash plm.sh
# Use PCG utility to populate with lipids
bash pcg.sh
This creates system.gro, which can be used for molecular dynamics simulation or visualization using e.g. Mosaic/VMD.
Equilibration#
Gromacs [7] settings for Martini [8] model equilibration are available from ownCloud in the ts2cg folder:
bash equilibrate.sh
This performs energy minimization which can be run on a standard laptop. The final equilibration step should be run in an HPC environment (see eq/equilibrate.sbatch for an example).
Conclusion#
You’ve now completed the entire workflow for analyzing IAV virus-like particles—from tomogram segmentation to creating a detailed molecular model. This model can serve as a foundation for structural analysis or as a starting system for molecular simulations.