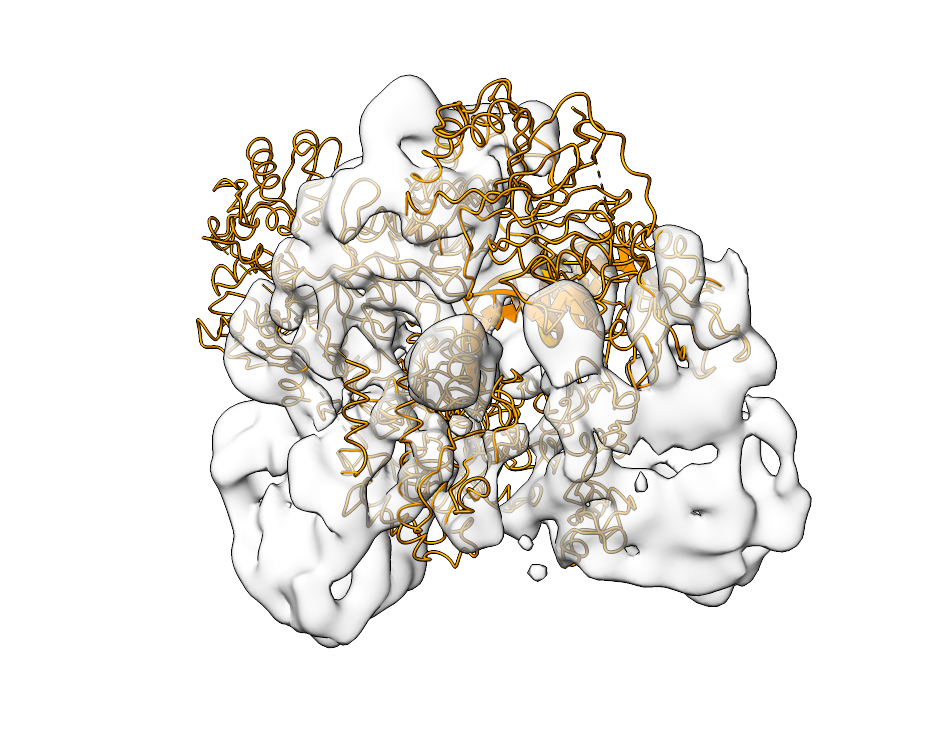
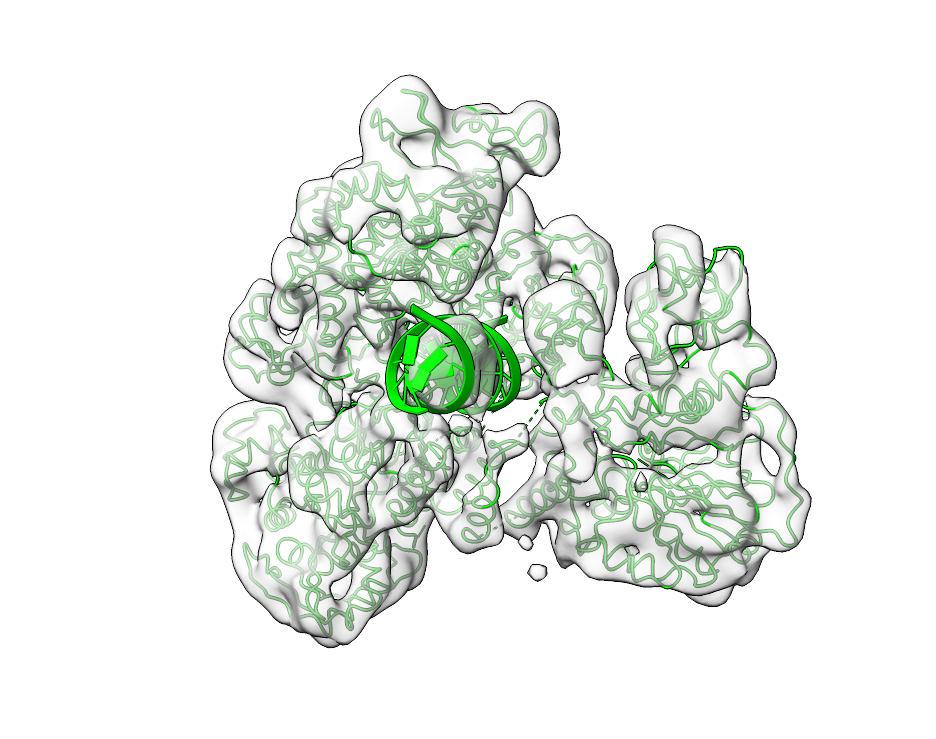
Fitting Atomic Structures#
In the following we will use template matching to recover the correct orientation of an atomic structure in a density map.
To demonstrate the procedure, we will use EMD-0244 and the associated fitted structure 6HMS. You can download both from EMDB or from the command line
wget https://ftp.ebi.ac.uk/pub/databases/emdb/structures/EMD-0244/map/emd_0244.map.gz
wget https://files.rcsb.org/download/6HMS.pdb
Since 6HMS is already in the correct orientation, we will first simulate a translation and rotation of the structure
from tme import Structure
from tme.rotations import get_rotation_matrices
rotation_matrix = get_rotation_matrices(40)[32].T
structure = Structure.from_file("6HMS.pdb")
structure_mod = structure.rigid_transform(
rotation_matrix = rotation_matrix,
translation = (-15, 10, 0)
)
structure_mod.to_file("6HMS_mod.pdb")
The following will fit the newly created structure 6HMS_mod.pdb into the density map to recover the correct orientation .. code-block:: bash
- match_template.py
-m emd_0244.map.gz -i 6HMS_mod.pdb -n 4 -a 40 –centering -o output.pickle
The orientation with highest score, which in our case is the initial correct orientation, can be obtained from the created pickle files like so
postprocess.py \
--input-file output.pickle \
--num-peaks 1 \
--output-format alignment \
--output-prefix 6HMS_fit
The postprocessing tool offers a range of additional features to enable subvoxel precision such as peak oversampling via --peak-oversampling and local optimization via the --local-optimization flag. The output of fitting is shown below. The left side shows the map and 6HMS_mod.pdb, the right side the output of pytme.